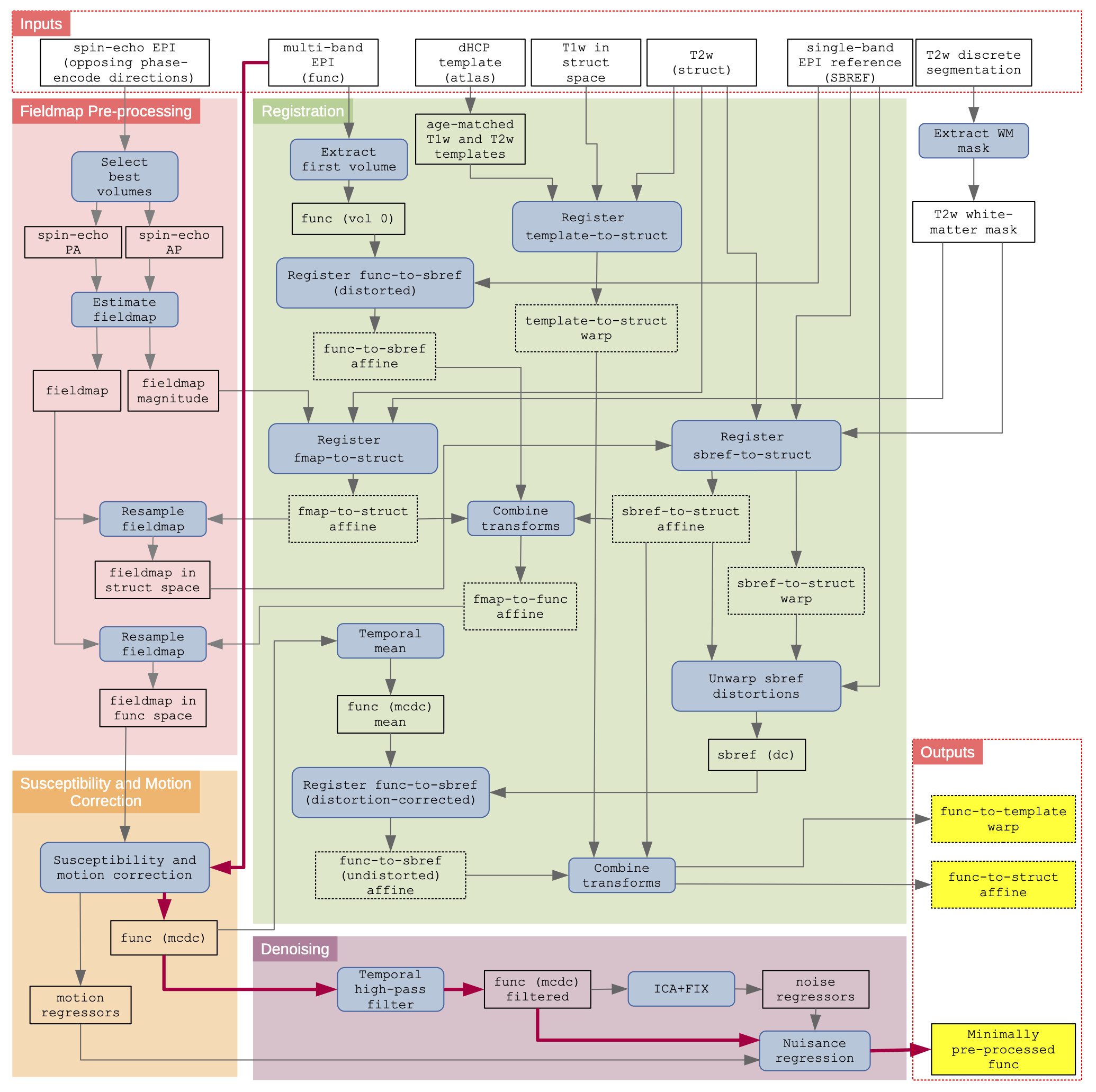
Functional (fMRI) Pipeline
Inputs
From reconstruction pipeline: rawdata/sub-{subid}/ses-{sesid}
| Description | Filename |
|---|---|
| Resting fMRI | func/sub-{subid}_ses-{sesid}_run-{seqnum}_task-rest_bold.nii |
| Single-band Ref | func/sub-{subid}_ses-{sesid}_run-{seqnum}_task-rest_sbref.nii |
| 4D Spin Echo EPI with different phase encode directions (for topup fieldmap estimation) | fmap/sub-{subid}_ses-{sesid}_run-{seqnum}_epi.nii |
| Dual echo-time (magnitude) | fmap/sub-{subid}_ses-{sesid}_run-{seqnum}_magnitude.nii |
| Dual echo-time field-map in (Hz) - filtered and smoothed | fmap/sub-{subid}_ses-{sesid}_run-{seqnum}_rec-filtered_fieldmap.nii |
From structural pipeline: derivatives/dhcp_anat_pipeline/sub-{subid}/ses-{sesid}
| Description | Filename |
|---|---|
| T2 weighted, bias corrected image | anat/sub-{subid}_ses-{sesid}_desc-restore_T2w.nii.gz |
| Draw-EM tissue segmentation (9 labels) | anat/sub-{subid}_ses-{sesid}_desc-drawem9_dseg.nii.gz |
| Left/Right white surface | anat/sub-{subid}_ses-{sesid}_hemi-{hemi}_wm.surf.gii |
| Left/Right pial surface | anat/sub-{subid}_ses-{sesid}_hemi-{hemi}_pial.surf.gii |
Outputs
The primary outputs of the fMRI preprocessing pipeline are:
Path: derivatives/dhcp_fmri_pipeline/sub-{subid}/ses-{sesid}
| Description | Filename |
|---|---|
| Multi-band EPI, distortion corrected, motion corrected, FIX denoised, 4D image | func/sub-{subid}_ses-{sesid}_task-rest_desc-preproc_bold.nii.gz |
| Warp from functional space to the extended dHCP 40-week template space | xfm/sub-{subid}_ses-{sesid}_from-bold_to-extdhcp40wk_mode-image.nii.gz |
The complete list of outputs and QC reports generated by the fMRI pipeline are listed in the Functional pipeline section of the directory structure summary.
Pipeline
Version 1.2.0 of the dHCP neonatal fMRI pipeline was used for release 3 and can be downloaded here: https://git.fmrib.ox.ac.uk/seanf/dhcp-neonatal-fmri-pipeline/-/tree/1.2.0
The preprocessing steps implemented in the pipeline are as follows:
-
Prepare fieldmaps for correction of susceptibility distortions
-
Estimate field map from the two “best” spin-echo volumes (1 per phase-encode direction) using FSL TOPUP1. “Best” is defined by smoothness in the z-dimension (stdev of the slice-to-slice difference in the z-dimension)
-
If visual inspection indicates that the spin-echo has significant motion contamination then use the dual-echo-time-derived fieldmap instead of the spin-echo-derived fieldmap
-
-
Registration
-
Boundary-based registration (BBR, FSL FLIRT3) of the fieldmap magnitude to the T2w structural
-
Boundary-based registration (BBR, FSL FLIRT3) of the sbref to the T2 structural incorporating field map-based distortion correction of the sbref
-
Linear registration (6-dof, corratio, FSL FLIRT3) of the first volume of the functional multiband EPI to the sbref
-
After susceptibility and motion correction (see below), linear registration (6-dof, corratio, FSL FLIRT3) of the temporal mean of the motion and distortion corrected functional multiband EPI to the distortion corrected sbref
-
Nonlinear diffeomorphic multimodal registration of the age-matched T2w and GM/WM tissue probability from the dHCP volumetric atlas7 to the subjects T2w and GM/WM tissue probability using ANTs SyN6. The GM/WM probability for each subject was calculated as the partial volume estimate from a tool called Toblerone. We have augmented the dHCP volumetric atlas with week-to-week nonlinear transforms estimated using a diffeomorphic T2w registration (ANTs SyN). The appropriate transforms are then combined to yield a (40 week) template-to-structural transform
-
From these primary registrations the following composite transforms are calculated:
-
Fieldmap to native functional
-
Motion and distortion corrected functional to 40-week template from the dHCP volumetric atlas
-
-
-
Susceptibility and motion correction
- Slice-to-volume motion correction and motion-by-susceptibility correction is performed using FSL EDDY2
-
ICA Denoising
-
Temporal high-pass filter (150s high-pass cutoff) and ICA denoising using FSL FIX4, pre-trained with manually-labelled data from 50 dHCP neonatal subjects, to identify artefactual ICs (accuracy: median TPR=98%, median TNR=96%). The ICA dimensionality was capped at 600 ICs.
-
Noise ICs and motion parameters regressed from motion and distortion corrected functional multiband EPI.
-
Schematic of the dHCP fMRI neonatal pre-processing pipeline: The schematic is segregated into the 4 main conceptual processing stages by coloured background; fieldmap pre-processing (red), susceptibility and motion correction (orange), registration (green), and denoising (purple). Inputs to the pipeline are grouped in the top row, and the main pipeline outputs are grouped in the lower right. Blue filled rectangles with rounded corners indicate processing steps, whilst black rectangles (with no fill) represent data. The critical path is denoted by magenta connector arrows. (dc) = distortion corrected; (mcdc) = motion and distortion corrected.
Quality Control/Assurance
-
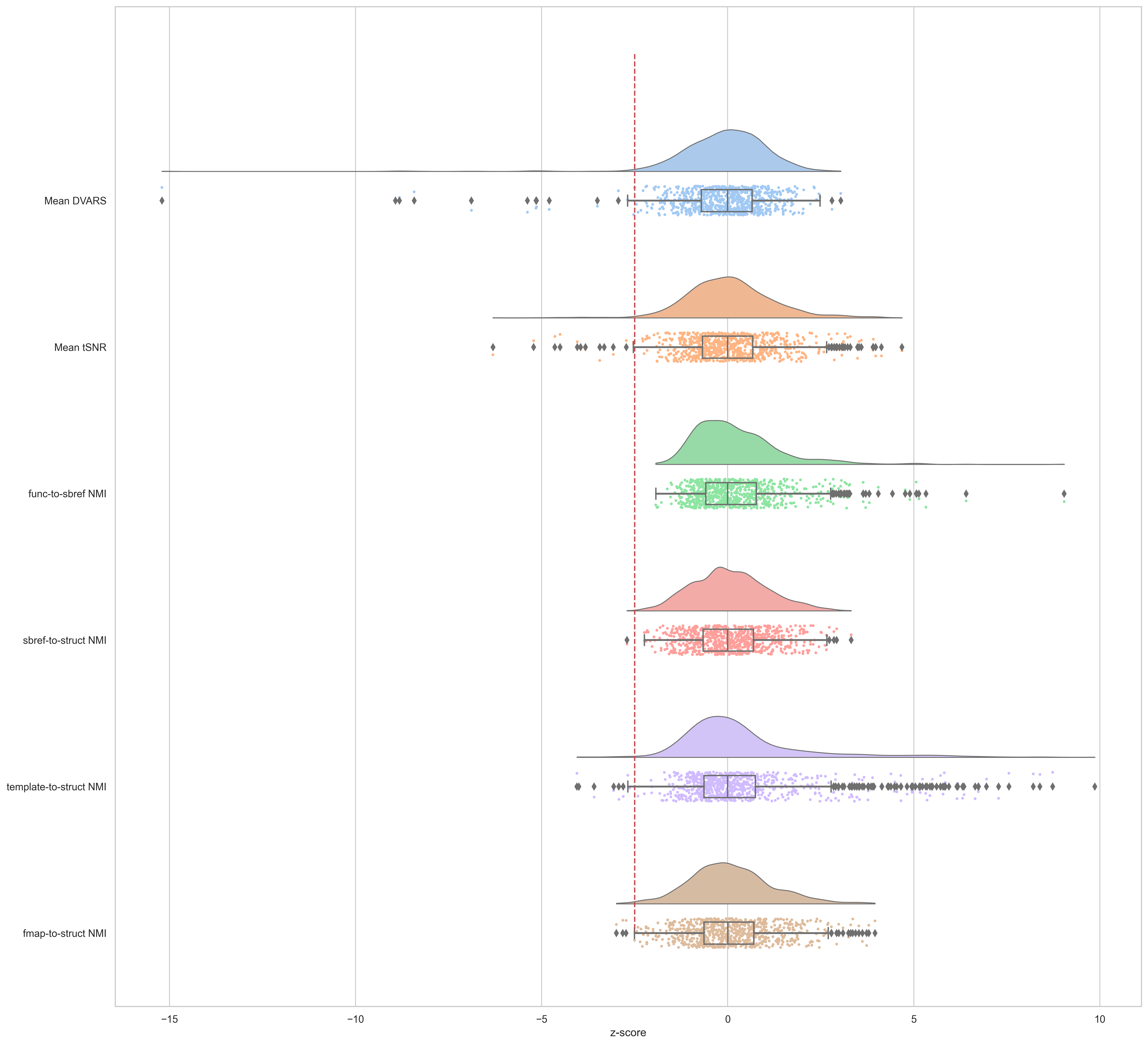
Numerous quality assurance metrics are calculated during the pre-processing. Six of these are specifically compared against the population distribution to flag outliers for manual inspection and potential exclusion:
-
Mean DVARS5 of the ICA denoised functional EPI; provides a snapshot of output signal quality
-
Mean tSNR of the ICA denoised functional EPI; provides a snapshot of output signal quality
-
Normalised mutual information of the source (moving) image, re-sampled to reference space, and the reference (fixed) image, for each of the primary registrations:
-
Fieldmap to structural T2w
-
Native functional to sbref
-
Motion and distortion corrected functional to sbref
-
Sbref to structural T2w
-
Age-matched atlas template T2w to native structural T2w
-
-
-
All QA measures were converted to Z-scores and flipped as necessary so that positive z-scores are good and negative bad. Subject/sessions with a z-score < -2.5 on any QC metric were flagged for further inspection.
-
All QC metrics are available in the
combined.tsvspreadsheet in the supplementary.
QC Scores: 28 subject/sessions were flagged for futher inspection (see figure). All flagged subject/sessions are included in the release.
How to cite
Detailed instructions on how to cite can be found here: http://www.developingconnectome.org/how-to-cite/
Primary citation for the fMRI pipeline:
Fitzgibbon, SP, Harrison, SJ, Jenkinson, M, Baxter, L, Robinson, EC, Bastiani, M, Bozek, J, Karolis, V, Cordero Grande, L, Price, AN, Hughes, E, Makropoulos, A, Passerat-Palmbach, J, Schuh, A, Gao, J, Farahibozorg, S, O’Muircheartaigh, J, Ciarrusta, J, O’Keeffe, C, Brandon, J, Arichi, T, Rueckert, D, Hajnal, JV, Edwards, AD, Smith, SM, *Duff, E, *Andersson, J The developing Human Connectome Project automated functional processing framework for neonates., NeuroImage (2020), 223: 117303, 2020. DOI: 10.1016/j.neuroimage.2020.117303 *Authors contributed equally.
@article {Fitzgibbon766030,
author = {Fitzgibbon, Sean P. and Harrison, Samuel J. and Jenkinson, Mark and Baxter, Luke and Robinson, Emma C. and Bastiani, Matteo and Bozek, Jelena and Karolis, Vyacheslav and Grande, Lucilio Cordero and Price, Anthony N. and Hughes, Emer and Makropoulos, Antonios and Passerat-Palmbach, Jonathan and Schuh, Andreas and Gao, Jianliang and Farahibozorg, Seyedeh-Rezvan and O{\textquoteright}Muircheartaigh, Jonathan and Ciarrusta, Judit and O{\textquoteright}Keeffe, Camilla and Brandon, Jakki and Arichi, Tomoki and Rueckert, Daniel and Hajnal, Joseph V. and Edwards, A. David and Smith, Stephen M. and Duff, Eugene and Andersson, Jesper},
title = {The developing Human Connectome Project (dHCP) automated resting-state functional processing framework for newborn infants},
elocation-id = {117303},
year = {2020},
doi = {10.1016/j.neuroimage.2020.117303},
publisher = {Elsevier},
URL = {https://doi.org/10.1016/j.neuroimage.2020.117303},
eprint = {https://www.sciencedirect.com/science/article/pii/S1053811920307898/pdfft?md5=18806cf190a26f783de4bef456fe28b6&pid=1-s2.0-S1053811920307898-main.pdf},
journal = {NeuroImage}
}
References
-
Andersson, J. L., Skare, S. and Ashburner, J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging Neuroimage (2003), 20: 870-888. DOI: 10.1016/S1053-8119(03)00336-7
-
Andersson, J. L. and Sotiropoulos, S. N. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging Neuroimage (2016), 125: 1063-1078. DOI: 10.1016/j.neuroimage.2015.10.019
-
Jenkinson, M., and Smith, S. A global optimisation method for robust affine registration of brain images Medical image analysis (2001), 5(2): 143–156. DOI: 10.1016/S1361-8415(01)00036-6
-
Salimi-Khorshidi, G., Douad, G, Beckman, C. F., Glasser, M. F., Griffanti, L., and Smith, S. M. Automatic denoising of functional MRI data: combining independent component analysis and hierarchical fusion of classifiers NeuroImage (2014), 90: 449–468. DOI: 10.1016/j.neuroimage.2013.11.046
-
Power, J. D., Barnes, K. A., Snyder, A. Z., Schlaggar, B. L., and Petersen, S. E. Spurious but Systematic Correlations in Functional Connectivity MRI Networks Arise from Subject Motion NeuroImage (2012), 59(3): 2142–54. DOI: 10.1016/j.neuroimage.2011.10.018
-
Avants, B.B., Epstein, C.L., Grossman, M., Gee, J.C. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. (2008), 12, 26–41. DOI: 10.1016/j.media.2007.06.004
-
Andreas Schuh, Antonios Makropoulos, Emma C. Robinson, Lucilio Cordero-Grande, Emer Hughes, Jana Hutter, Anthony N. Price, Maria Murgasova, Rui Pedro A. G. Teixeira, Nora Tusor, Johannes Steinweg, Suresh Victor, Mary A. Rutherford, Joseph V. Hajnal, A. David Edwards, and Daniel Rueckert. Unbiased construction of a temporally consistent morphological atlas of neonatal brain development, bioRxiv, 2018. DOI: 10.1101/251512